Chelicerate phylogenomics
Chelicerata constitutes a diverse branch of arthropods that includes such lineages as spiders, mites, and horseshoe crabs.
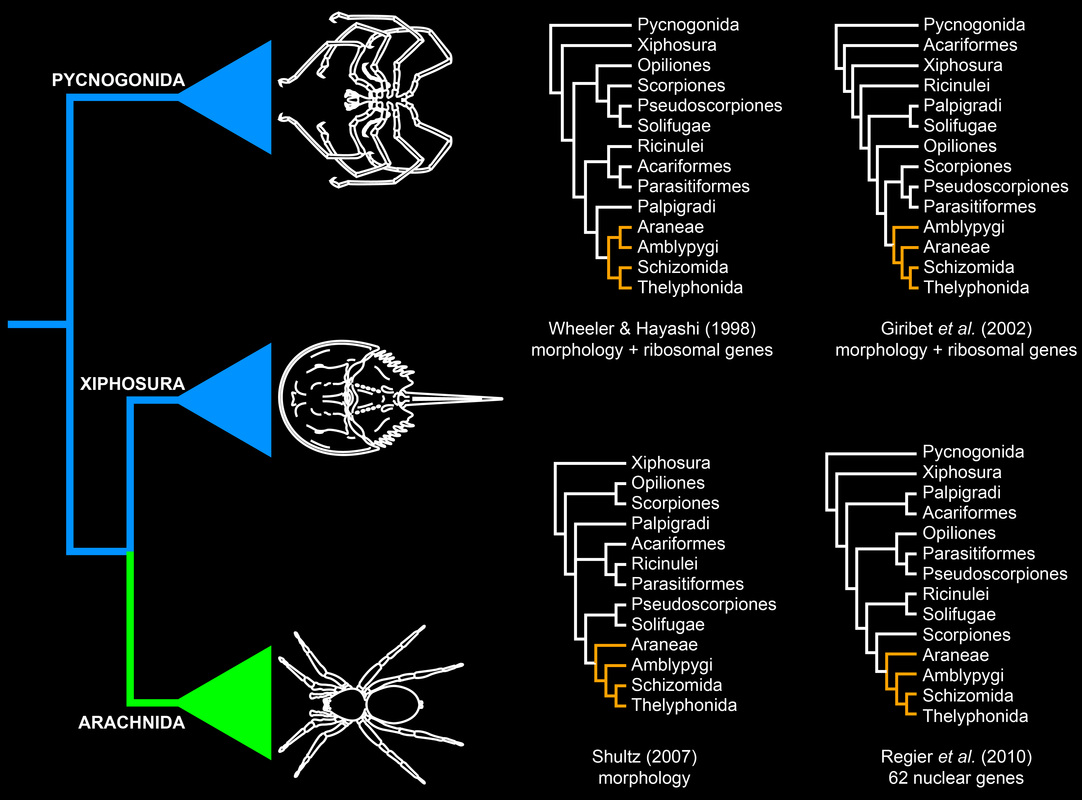
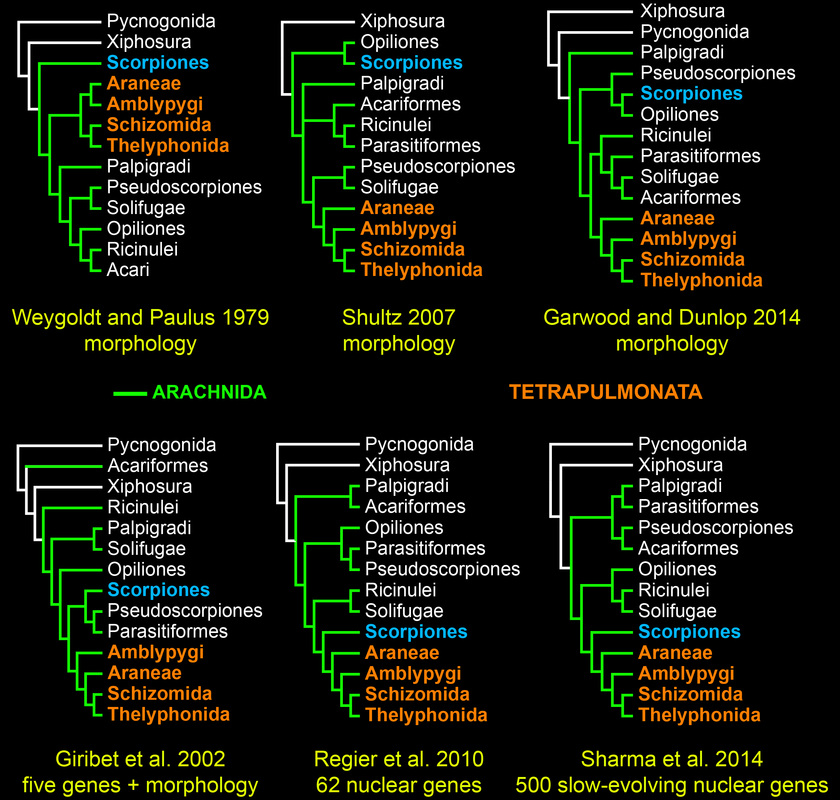
Chelicerate phylogeny has long been a recalcitrant systematic problem. Various datasets and approaches have yielded discordant, poorly supported basal topologies. Molecular sequence data in particular have difficulty with recovering the monophyly of Arachnida (the terrestrial chelicerates) with support. In the figure below, the traditional understanding of chelicerate relationships is depicted on the left.
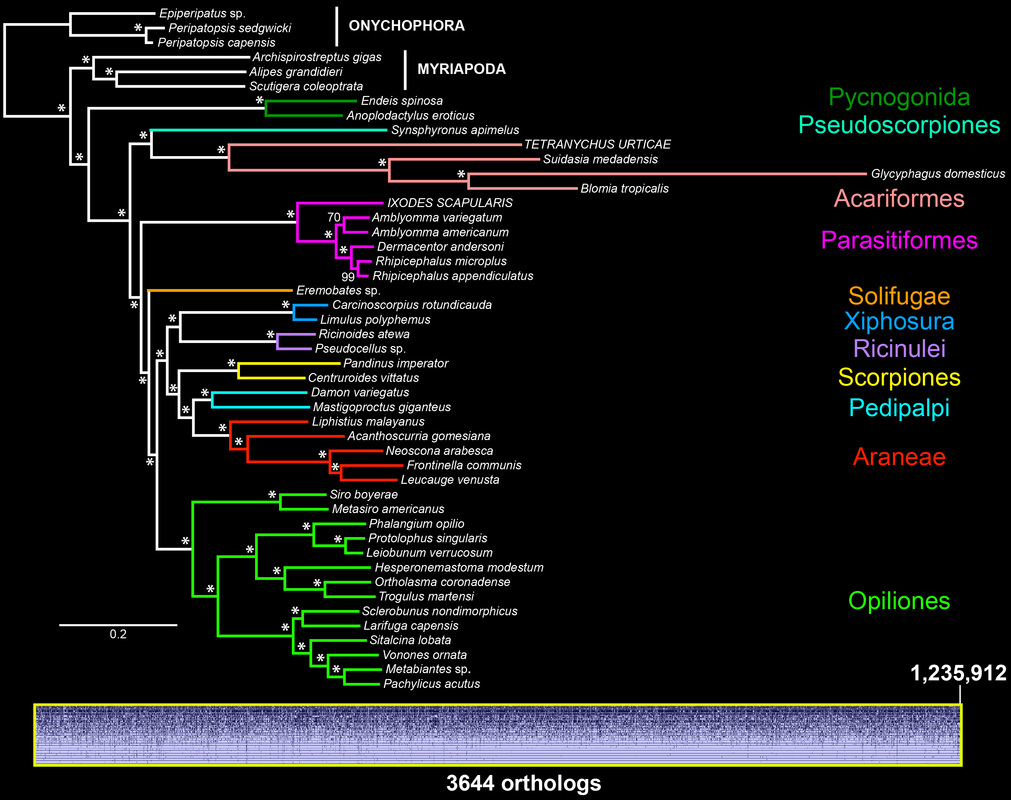
We inferred chelicerate phylogeny using deep sequencing technology and a fully computational approach to orthology prediction, assembling a supermatric of >3600 orthologs. Not surprisingly, the resulting topology (below) yielded the non-monophyly of Arachnida, a result driven by long branch attraction (LBA) artifacts in problematic orders like mites (Acariformes), ticks (Parasitiformes), and pseudoscorpions. Note the placement of Xiphosura within the arachnids.
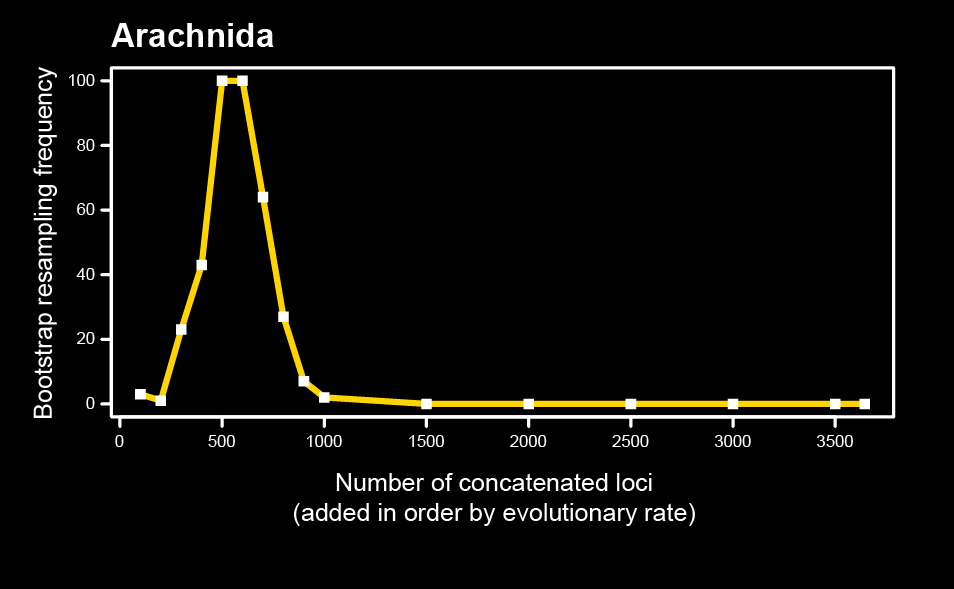
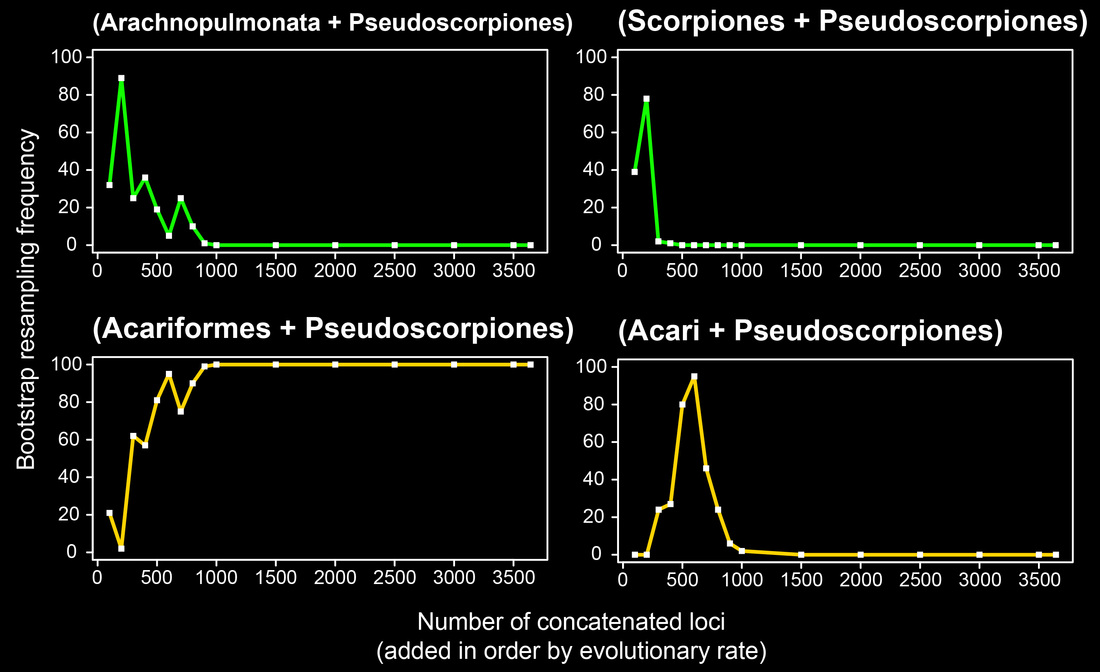
We examined several candidate explanatory variables driving this phenomenon, such as matrix completeness and algorithmic approach. The most powerful of these variables was discovered to be evolutionary rate. We found that concatenating orthologs in order of evolutionary rate (starting with the slowest evolving) recovered support for key relationships consistent with morphology. In the panel below, the localized phylogenetic support profile is shown for Arachnida.
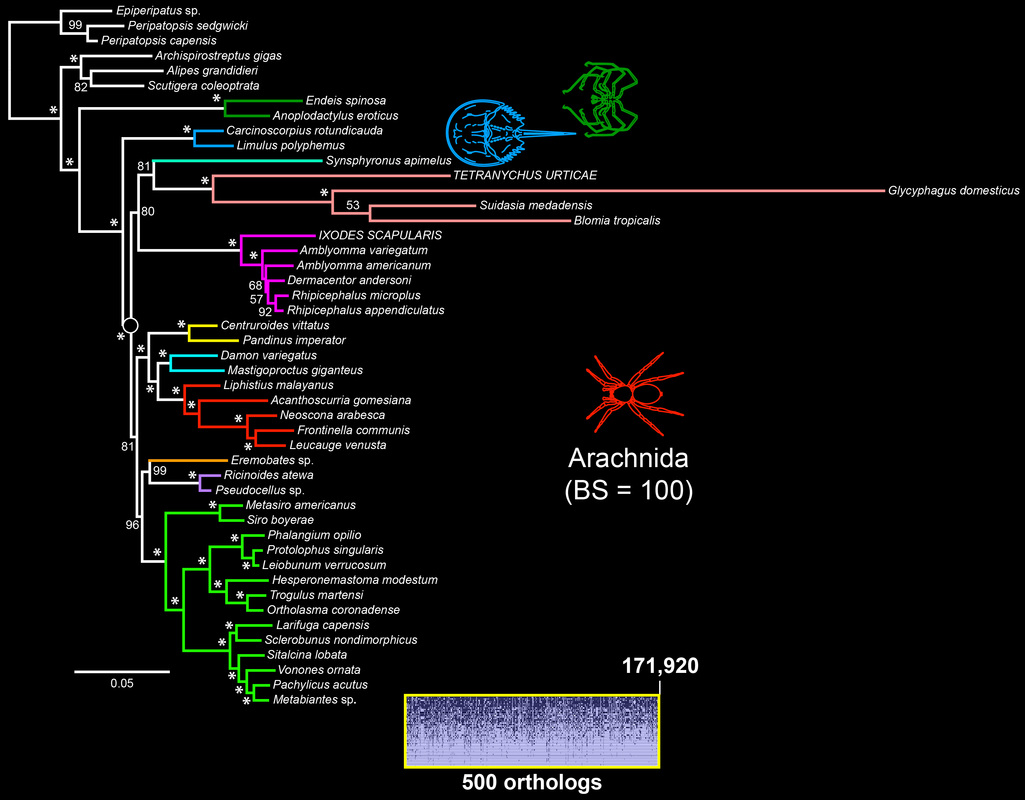
The tree topology based on the 500 slowest-evolving orthologs recovered for the first time the traditional basal topology of Chelicerata with full nodal support in a molecular phylogenetic study.
However, we also showed that localized phylogenetic support is not a sufficient condition of phylogenetic accuracy. For long branch lineages like Pseudoscorpiones, multiple mutually exclusive hypotheses are supported by various groups of orthologs. In the figure below, the green curves indicate nested hypotheses consistent with morphology, and the yellow curves a second pair of nested hypotheses that are inconsistent with morphology. All four receive some degree of support from various partitions.
While chelicerate relationships remain controversial (see Garwood and Dunlop 2014), the recovery of scorpions as sister group to the tetrapulmonates (spider and their allies) with high nodal support in the majority of our analyses is consistent with a single origin of the arachnid book lung.