Genomics
Genomics permeates nearly every aspect of the lab's operations. Both the phylogeneticists and the developmental biologists in the lab establish and use genomic toolkits to test evolutionary hypotheses and facilitate mechanistic understanding of key transitions in invertebrate biology.
Examples of recent projects
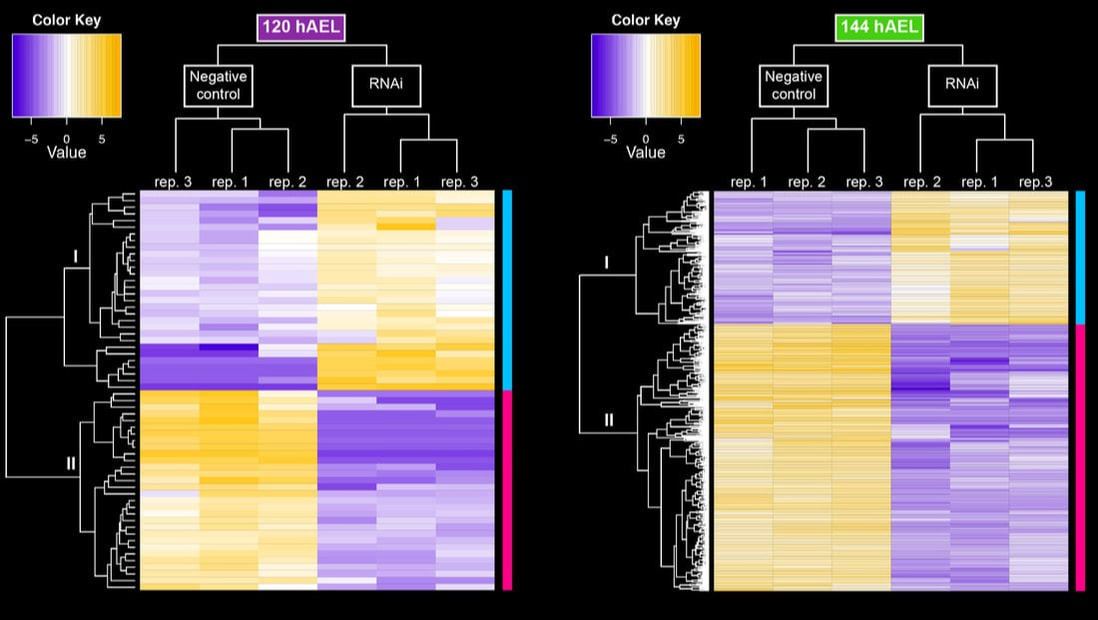
Leveraging the relatively recent publication of a high-quality genome for the house spider, we have used RNA-Seq approaches in tandem with differential gene expression analyses (DGE) to characterize complex RNAi phenotypes. This approach provides a systemic view of the effect of gene disruption on developmental processes, such as segmentation or limb development.
Examples of recent projects
Leveraging the relatively recent publication of a high-quality genome for the house spider, we have used RNA-Seq approaches in tandem with differential gene expression analyses (DGE) to characterize complex RNAi phenotypes. This approach provides a systemic view of the effect of gene disruption on developmental processes, such as segmentation or limb development.
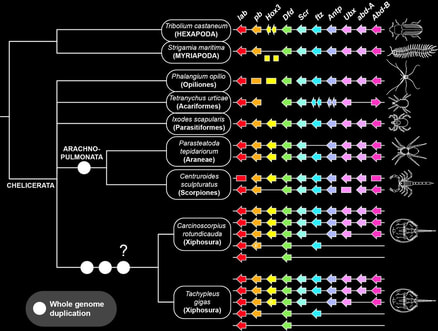
Beyond established models, the lab is actively pursuing in generating genomic resources for poorly studied arachnid groups, such as whip spiders, harvestmen, pseudoscorpions, and sea spiders. Recently, we published the first harvestman genome, in tandem with functional investigation of leg patterning and elongation mechanisms in Phalangium opilio (we made daddy-long-legs with short legs!).
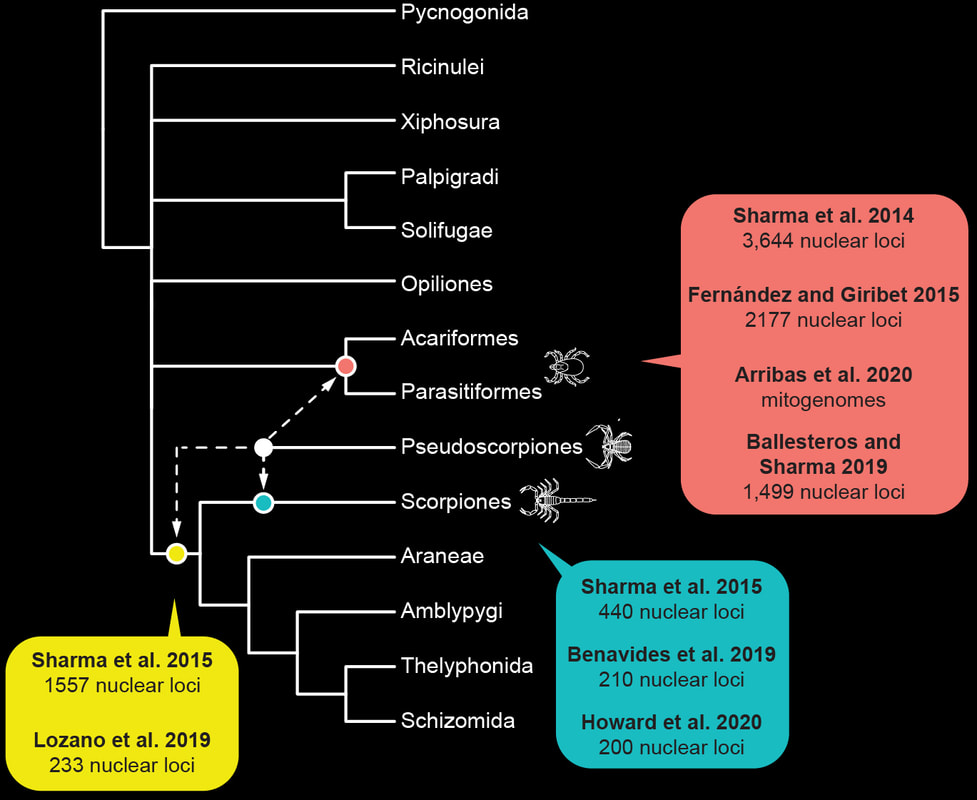
Genomics is instrumental to resolving difficult phylogenetic questions in Chelicerata. In a collaboration with the Zeh Lab, we recently published the first developmental genetic resources, as well as the first draft genome, of pseudoscorpions. These datasets were used to establish whether pseudoscorpions are part of Arachnopulmonata (tetrapulmonates and scorpions, groups that share an ancient genome duplication) or closely related to the acarine orders (mites and ticks, which have unduplicated genomes).
We were able to show that pseudoscorpions have duplicated genomes and are clearly part of Arachnopulmonata. This discovery has aided in benchmarking methods for overcoming phylogenetic artifacts like long branch attraction.