Comparative developmental genetics and evolutionary developmental biology
Much of the lab's operations are dedicated to comparative developmental genetics or "evo-devo". This facet of the lab aims to unravel the genetic mechanisms that underpin macroevolutionary transitions in the animal tree of life, such as the origins of key innovations and the genetic basis for body plan patterning. Much of this work requires establishing de novo resources and emerging model systems from the ground up.
The lab has collectively established five new species for the study of arachnid comparative development and evo-devo: Phalangium opilio (harvestman), Centruroides sculpturatus (bark scorpion), Conicochernes crassus (pseudoscorpion), Phrynus marginemaculatus (whip spider), and Aphnopelma hentzi (Texas brown tarantula). Some projects also make use of a well-established system, Parasteatoda tepidariorum (cobweb spider). Learn more about our animals in The Zoo.
Snapshots from recent projects
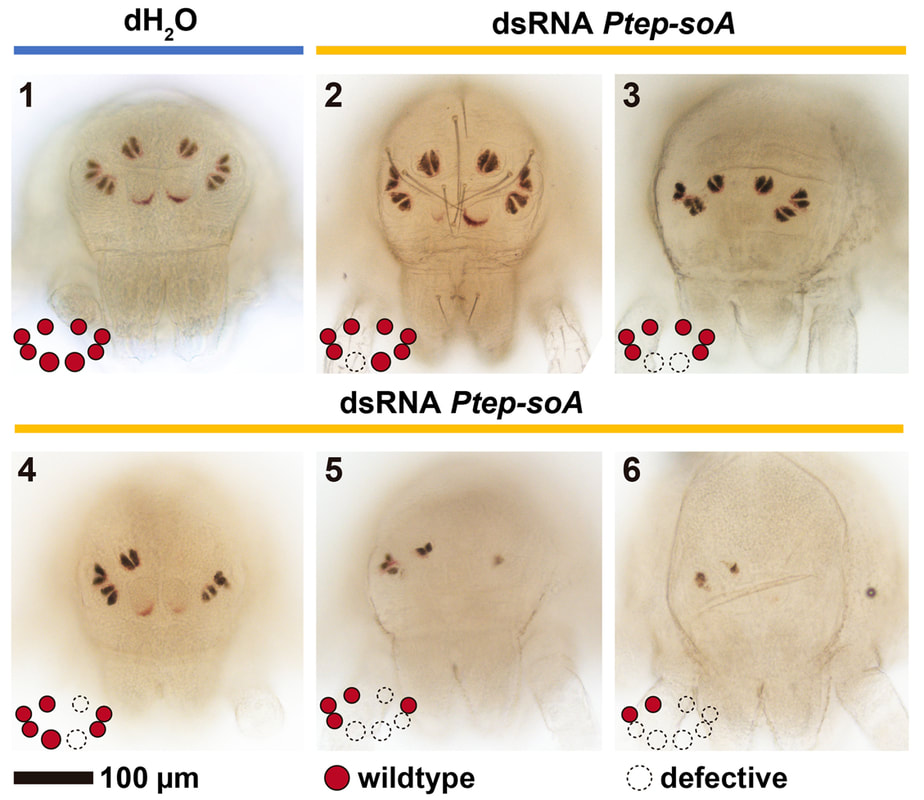
How spiders make their eyes: We recently showed that one of the two copies of sine oculis is required for both median and lateral eye development in the spider Parasteatoda tepidariorum. This represents the first functional genetic data point for eye patterning in any arachnid species.
The lab has collectively established five new species for the study of arachnid comparative development and evo-devo: Phalangium opilio (harvestman), Centruroides sculpturatus (bark scorpion), Conicochernes crassus (pseudoscorpion), Phrynus marginemaculatus (whip spider), and Aphnopelma hentzi (Texas brown tarantula). Some projects also make use of a well-established system, Parasteatoda tepidariorum (cobweb spider). Learn more about our animals in The Zoo.
Snapshots from recent projects
How spiders make their eyes: We recently showed that one of the two copies of sine oculis is required for both median and lateral eye development in the spider Parasteatoda tepidariorum. This represents the first functional genetic data point for eye patterning in any arachnid species.
How daddy-long-legs make their long legs: Recently, we published the first harvestman genome, in tandem with functional investigation of leg patterning and elongation mechanisms in Phalangium opilio (we made a "daddy-short-legs"!).
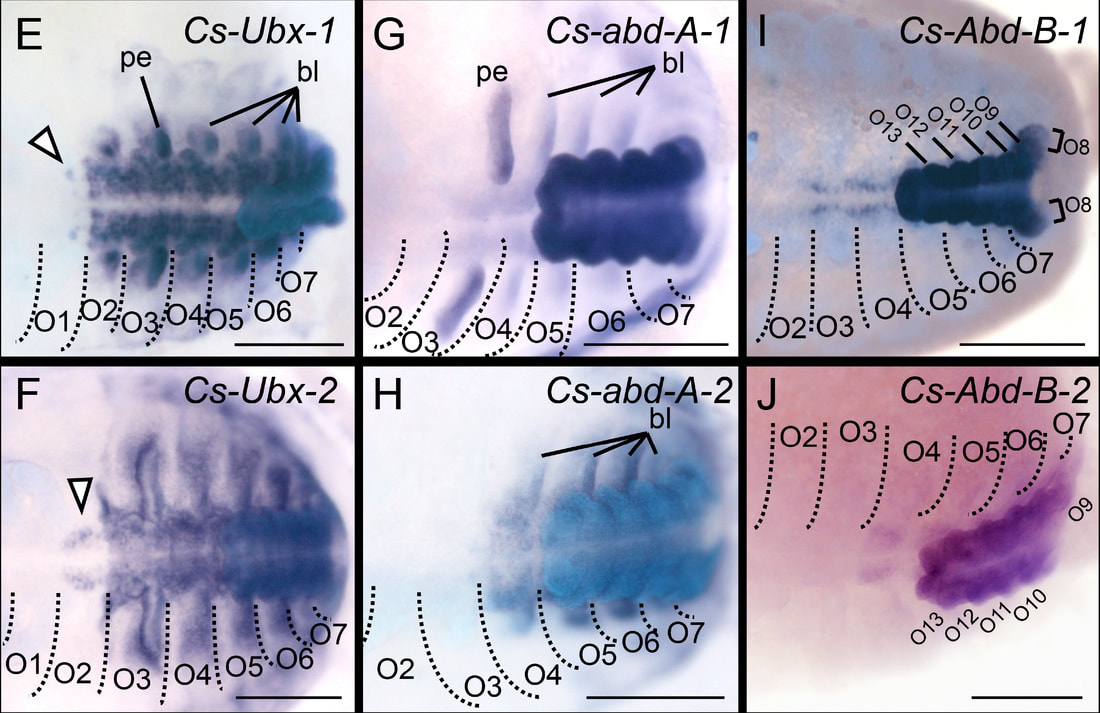
How scorpions make their tails: Applying transcriptomics to survey developing embryos of scorpions, we discovered that nearly all scorpion Hox genes have been duplicated. Expression patterns of the last four pairs of Hox genes in scorpion embryos demonstrate unique anterior expression boundaries, suggesting that the scorpion bauplan was achieved by neofunctionalization of Hox gene paralogs.
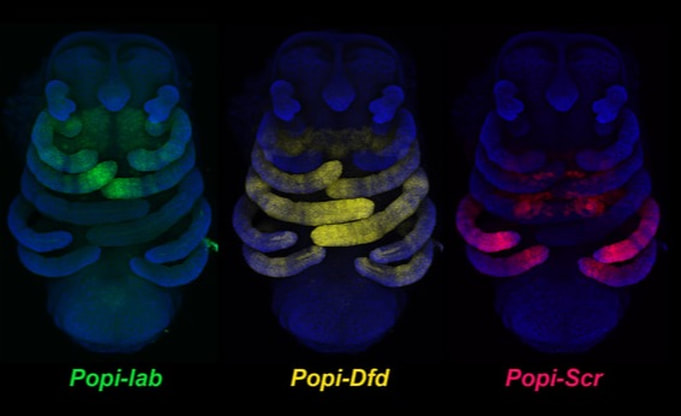
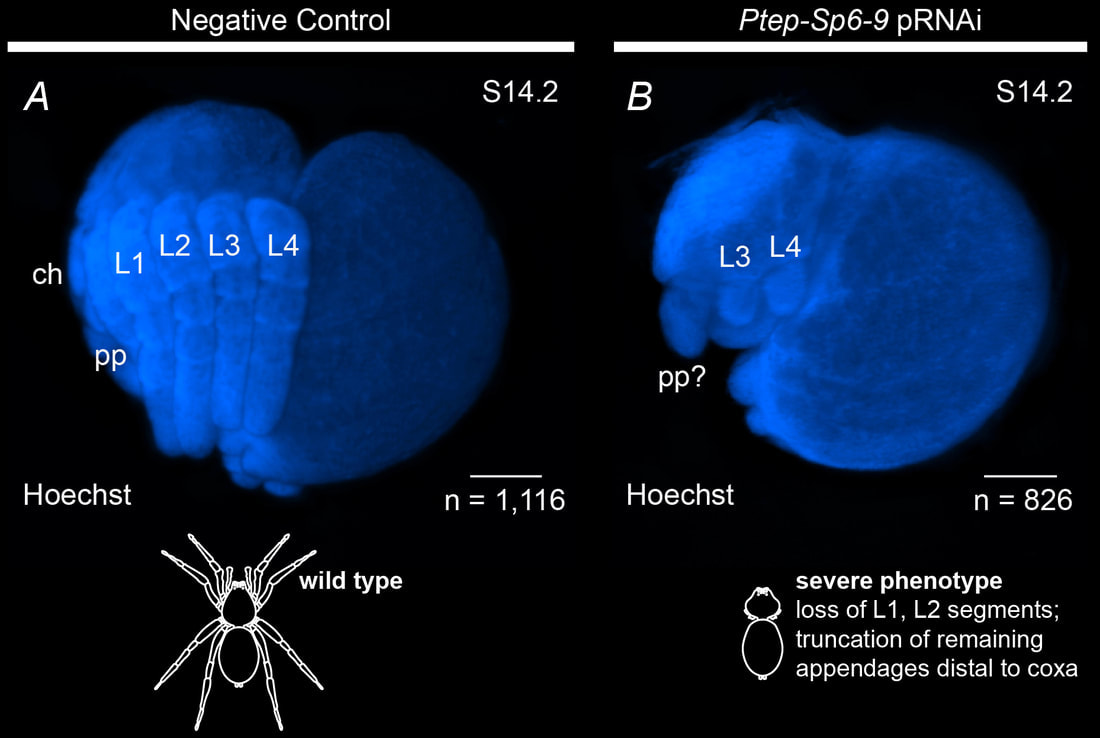
How arachnids make their head segments: We used a combination of genomics, developmental genetics, and comparative gene expression surveys to discover that arachnids have coopted leg-patterning genes to build their head segments.
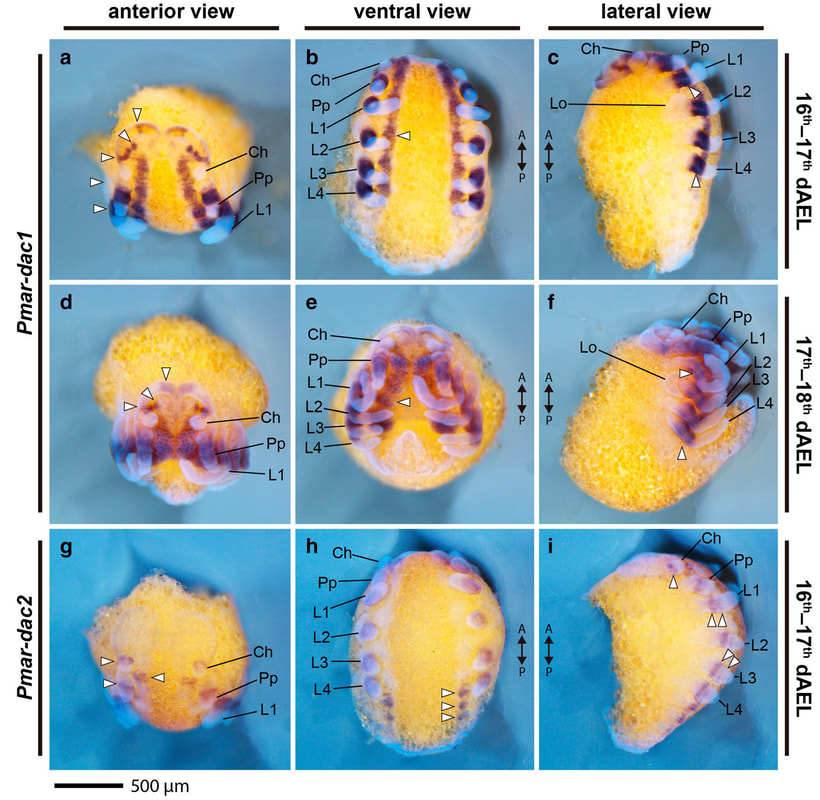
How whip spiders deploy duplicated gene copies: We resurrected the studies of Peter Weygoldt on the whip spider species Phrynus marginemaculatus to investigate how this arachnopulmonate expresses duplicated copies of ancient transcription factors. Our goal is to understand how the first walking leg of this species was modified into the antenniform "whip".