Scorpion venom is a mixture of diverse components, traditionally divided into two fractions: toxic and non-toxic. The toxic fraction includes all those peptides affecting ion channels; the non-toxic fraction includes enzymes, lipids, mucopolysaccharides, etc. While the toxic fraction has been the most studied, next sequencing generation, and the study of venom transcriptomes, have allowed us to explore the diversity of all components.
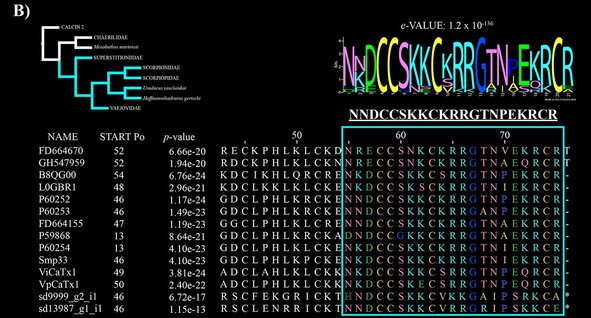
Using scorpion venom transcriptomes, I study, with Prof. Prashant Sharma and Dr. Ricardo Kriebel from the University of Wisconsin-Madison, and with Prof. Lourival Possani, from the Biotechnology Institute of UNAM in Mexico, the evolution of Calcins, which are peptides with high affinity to the ryanodine-sensitive calcium channel of the endoplasmic and sarcoplasmic reticula and skeletal and cardiac muscle (Santibáñez-López et al. 2016, Toxins 8, 367).
Using scorpion venom transcriptomes, I study, with Prof. Prashant Sharma and Dr. Ricardo Kriebel from the University of Wisconsin-Madison, and with Prof. Lourival Possani, from the Biotechnology Institute of UNAM in Mexico, the evolution of Calcins, which are peptides with high affinity to the ryanodine-sensitive calcium channel of the endoplasmic and sarcoplasmic reticula and skeletal and cardiac muscle (Santibáñez-López et al. 2016, Toxins 8, 367).
The phylogeny of these peptides is congruent with the phylogeny of scorpion families. In a previous work, we found the mature peptide of the calcins have this motif: XXXCCS(G)XXCXR(K)R(K)GXXXXXR(K)CX; where X represent variable amino acids (but see figure B below), and the letter in parenthesis represents an alternative to the previous amino acid.
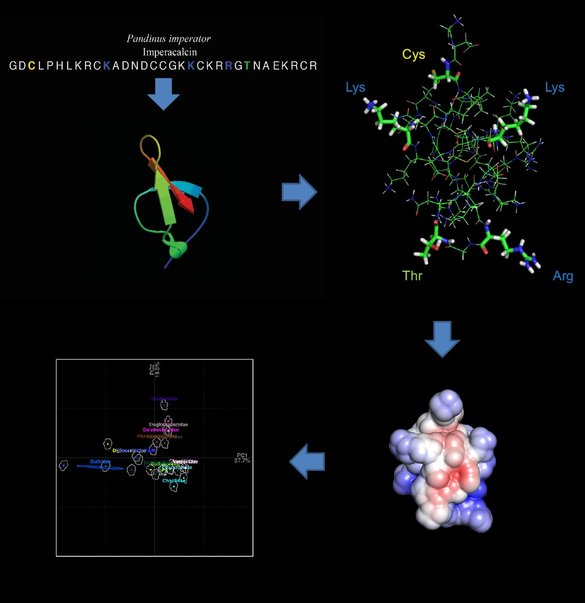
From there, we are studying the molecular evolution of these peptides, we want to understand the selective pressure (positive or negative) on amino acid sites, and what these changes reflects on the 3D structure of these peptides. Thus, we are modeling the structures of calcins recovered from more than 50 venom transcriptome analyses. We are projecting the biomolecular solvation using as a template the 3D model of maurocalcin, extracted from the venom of Scorpio maurus palmatus; and the Adaptive Poisson-Boltzmann Solver (APBS) package. Then, once we have all calcins modeled, we are projecting the variation of these 3D structures into the morphospace, using the position of certain amino acids as landmarks.